PiSITE tutorial
How to access each entry
The user can access each PiSITE entry in two different ways. The first way is accessing by a compiled list. PiSITE now provides the list of sociable proteins (transient hub-proteins), which have many binding states and partners. The lists contains links for entries, and the user can access them by just clicking the link. The other way to access PiSITE is search. PiSITE accepts both keywords and PDB ID for the search.
Searching PiSITE
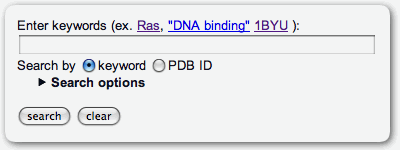
The keyword search for PiSITE entries can be available through the search form at the top page. This search ignore the case of letters in search keywords. Thus, 'RAS' and 'Ras' are dealt with same word. If the query contains multiple keywords separated with spaces or tabs, the AND search is performed. (If the query is 'dna binding', the query is dealt with 'dna AND binding') If user want to search by sentences, the sentence have to be quoted by double quotation, for example, "DNA binding protein". Notice that single quotation can not be used for this purpose. The PDB ID is aviailable as a keyword. If the PDB ID of a target protein is already known, then a rapid search can be available selecting the search by PDB ID radio button. The User can set the conditions for the search by the number of binding states, partners, and similar proteins. through search options. The search options are ordinary hided, and the user can access them by clicking the title.
Main view
Interaction viewer
The main view of the PiSITE is interaction viewer for each protein chain. This page mainly consists of three parts, title table, binding state viewer, and molecule viewer.
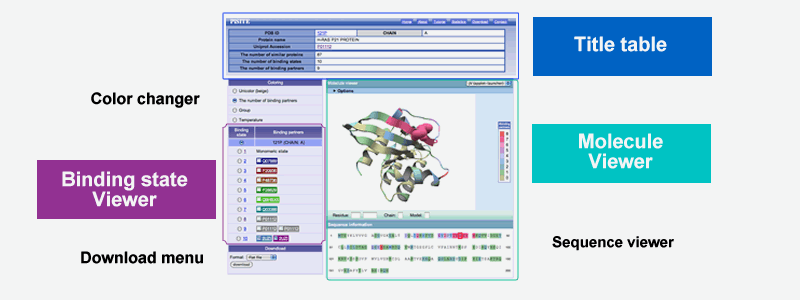
Title table
Title table shows the fundamental information of the entry including PDB ID and chain name, protein name and the accession number of external protein databases, if available. The table also includes the information of the number of same protein in PDB (the number of similar proteins) the number of binding states of the protein (Number of binding states) and the number of the class of the binding partner proteins (Number of binding partners)
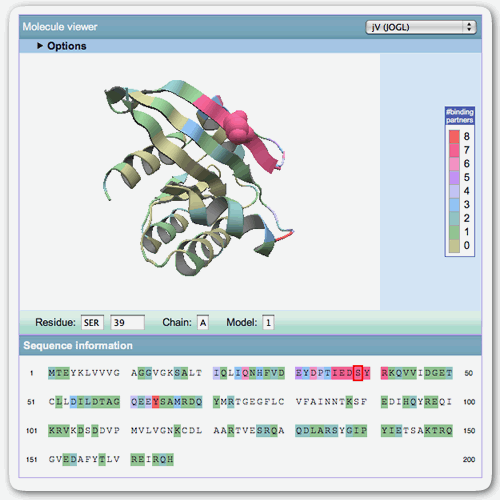
Molecule viewer
The molecule viewer shows the protein whose residues are colored by the number of binding partners by default. This picture insists the important interaction sites used for binding multiple partners.
The sequence of the protein chain is provided at lower part of moleculer viewer. Each letter of the sequence is clickable. The user can check the position of target residue by clicking the amino acid letter of the residue. The back-color of seqeunce is same in the 3D-picture.
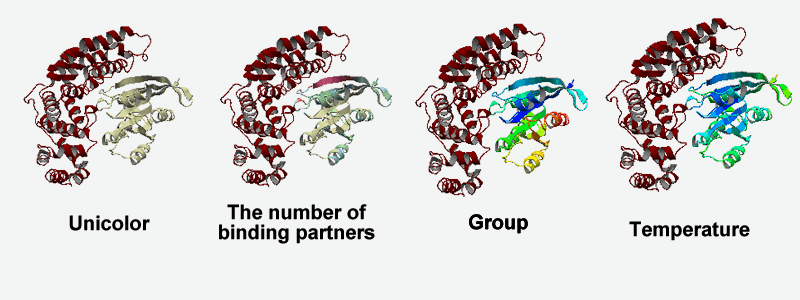
Color of the molecule can be changed through color changer at left-side. Now, four coloring schemes, unicolor, the number of binding partners, group and temperature are available.

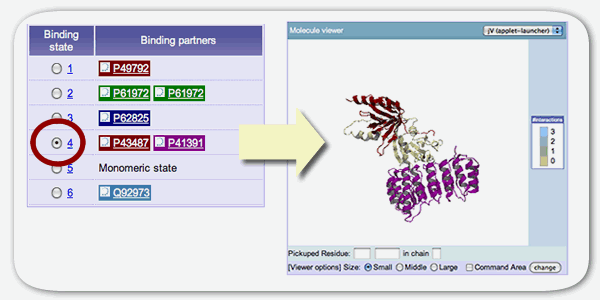
Binding state viewer
The binding state viewer shows the detail of each binding state of the protein including the information of binding partners. Each column corresponds to the each binding states and binding partners are diplayed in the accession number of UniProt or GenBank ID or PDB ID. The background color of the binding partner indicate the kind of the protein. The user can select left radio box to check the binding state at Molecule viewer. In this view, each protein chian is drawn by same color at the binding state viewer. The gray always indicates that binding partner is a same protein chain to the target protein chain.
Download
The binding information for each protein chain can be downloaded from the main view. Download menu is at the left bottom. The user can select the format of downloading data from XML and flat file. The detail of the flat file format can be available at download page. The download page can be accessed through header menu.
